home
 information
information  experimental design: replicates
experimental design: replicates
There are two main types of replicates that can be performed in a microarray experiment: biological and technical.
Biological Replicates:
- Preparing multiple independent samples for each treatment
- Allow for assessment of natural biological variance
Technical Replicates:
- Using more than one array to test a single sample
- Example: Multiple spots of the same probe, such as duplicate spotting
- Allow for assessment of measurement error
- Becoming less important as microarray technology matures
Replicate spotting is no longer of much value to the researcher as the variance between spots (especially when they are printed side-by-side) is extremely low. The most common (and perhaps useful in some cases) example of replicate arrays being used is in the case of reciprocal labelling (aka fluor-flips or dye-swaps).
Reciprocal Labelling:
In the days of two-colour microarray experiments, there were reports of strange and inconsistent data that seemed to be related to the fluorophore used to label a particular sample. Once data from controlled experiments was obtained, it became evident that certain RNAs seemed to label more efficiently with one flour or the other. This would often make it appear that a gene was being differentially regulated when it really was not. This dye bias was quite prevalent with some of the early direct labelling schemes.
To compensate and to isolate the effect, researchers found it useful to employ a technique which has been referred to as reciprocal labelling, dye-swaps or fluor-flips. The basis of this technique is that two hybridisations are performed for every pair of samples being analysed as shown in the following figure:
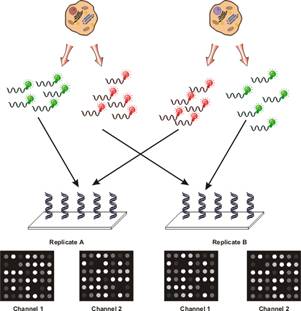
It was possible to isolate the effects of dye bias from true changes in gene expression. If a gene was found to be stronger in the green channel in the first hybridisation and in the red channel in the second hybridisation, this was likely due to an actual difference in abundance for that gene. If another gene was found to always have a higher signal in the green channel, this indicated that for some reason, the green fluor more efficiently labelled the mRNA for the particular gene.
What causes dye bias?
There has no been no conclusive answer. The leading suggestion is that due to the size and hydrophobicity differences of the fluorophores, they are incorporated with different efficiencies. In particular, it appears that reverse transcriptase (RT) is more tolerable of Cy3 than Cy5 (and their variants). As such, it would appear that Cy3 is incorporated with a generally higher efficiency. For certain mRNAs, particularly those with high secondary structure, Cy5 may be incorporated with lower efficiency. In general, however, it seems that for most mRNAs, there is no real difference between the two fluorophores.
It is suspected that Cy5 and its analogues lowers the overall processivity of the RT enzyme. If the RT enzyme hits a spot on an RNA molecule that has a particularly complex region of secondary structure AND at the same time needs to incorporate a Cy5 residue, it simply cannot proceed and falls off of the template. This theory is somewhat supported by the fact that newer variants of RT such as SuperScript III, which can be incubated at higher temperatures during labelling, seem to exhibit lower degrees of dye bias (the higher temperature is thought to further unfold any mRNA secondary structures).
Another theory is that RT has a hydrophobic binding domain, and that if a hydrophobic molecule binds to this domain, it inactivates the enzyme. Cy5 is more hydrophobic than Cy3, so it may inhibit the RT more frequently. However, this theory does not explain why the bias seems to be gene specific, and not simply a lower efficiency for one fluor over the other.
Can dye bias be eliminated?
An early strategy was to simply stop using two fluors and switch to a single channel design. While this eliminated the dye bias, it reduced the power of the technology, and in some cases increased costs because more arrays were required per experiment.
An alternate idea is the "indirect method", using an intermediate molecule, an aminoallyl, to label the DNA. The same molecule (aminoallyl-dUTP or aminoallyl-dCTP) is incorporated into each cDNA pool. The downstream chemical reaction to link the fluors to the cDNA is much more consistent between the two fluorophores, eliminating much of the bias.