home
 information
information  troubleshooting & help
troubleshooting & help  image quality
image quality
This collection of images illustrates various in-house array images. Does your image look like one of these? Click on the images to find out more about common problems and how to prevent lower quality images.

-
This is an excellent image! It is characterised by a range of spot intensities (dynamic range), minimal (if any) background, and minimal saturated spots. The spot morphology is close to perfect.

-
High background is the presence of signal on the areas of the array that are not spotted with nucleic acids. Most often caused by improper hybridisation or washing techniques.
Prevention: It is essential to prevent the hybridisation solution from drying onto the slide during hybridisation. Low humidity in the hyb chamber often results in high background (seen as streaks or bands around the perimeter of the coverslip). Hybridisation at increased temperatures (more than 42°C) can also cause background if the humidity is not high enough. Proper washing, as indicated in the standard protocols, is also critical. Be sure to agitate the slides occasionally during the washing process and rinse the slides with a non-SDS/low-salt solution. It is also important to dry the slides immediately after washing to prevent water droplets from drying.

-
Ghost spots are characteristic black spots on the array where the nucleic acid is printed. Often, ghost spots are obvious because the area surrounding the spot has higher background than usual. It appears that the spot of nucleic acid has actually repelled the labelled-cDNA or has become hydrophobic for some reason. The cause of ghost spots is unknown but sometimes occurs in older slides.
Prevention: It is difficult to prevent ghost spots but may occur less frequently if slides are stored in dry conditions (a desiccator is best) and used before they reach about 8-10 weeks old.

-
Donuts are spots that appear to have a hole in the middle. Donut spots are likely caused when the humidity is too low and the spotting solution dries too quickly on the array, causing the nucleic acids in the spotting solution to congregate around the perimeter of the spot.
Prevention: Avoid printing arrays when the humidity is too low (less than 40% RH).

-
Dumbbells and merged spots are also printing errors. As the name suggests, dumbbells are two spots that have merged together and look like a dumbbell in shape. Merged spots can form when the humidity is too high during printing. Merged spots can also be caused when the contact time between the printing pin and the array is too long, resulting in too much spotting solution being spotted down.
Prevention: Avoid printing arrays on very humid days (more than 60% RH). Optimize the contact time between the printing pins and the arrays.
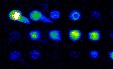
-
Comets are spots that have tails. These spots cause problems when it comes to quantification of the array since these spots are not circular. Quantification software like ArrayVision™ has an algorithm that can account for most irregular shaped spots. Comets are a printing error and are most often caused when the humidity is high. While precautions are taken to minimize humidity in the printing rooms, summers in Toronto can be very humid! Comets can also be caused when the outside of the printing pins are covered with spotting solution-nucleic acid.
Prevention: Avoid printing arrays on very humid days (more than 60% RH). Be sure to blot the printing pins on a glass surface (following each dip into the source plate) prior to printing on the arrays.

-
Scratched slides appear to have the spots scraped off the surface and causes problems when quantifying the array image. Scratches are often caused by scraping the array surface with forceps during the washing procedure. Scratches can also occur when removing a coverslip that has become dried onto the slide.
Prevention: Use caution when holding the slide with forceps during washing. Be sure to add enough hybridisation solution to slide and hybridisation box to prevent evaporation and subsequent drying.
- I am observing patches of background on my slide.
-
There are several causes of overall background found on the slide:
- Incomplete purification of labelling reaction - unincorporated Cy-dyes will stick to the arrays and give rise to fluorescent background upon scanning.
- Drying of the hybridisation mixture during overnight incubation causing the coverslip to stick to the arrays making it difficult to slide off during the washing process.
Remedy:- Follow our suggested protocol using CyScribe™ GFX™ columns (GE Healthcare) to remove all unincorporated dyes before hybridisation.
- Ensure a tight seal of the hybridisation chamber. When background is observed during prescanning of the microarray, consider rewashing it at either higher temperature or in a solution with a lower salt concentration to increase stringency of the washes. Be sure to save a high resolution scan prior to rewashing in case the rewash makes the image worse. Click here to read the Technical Note about rewashing arrays.
- I see signals from the 3X SSC spots when they should not hybridise to the labelled probe.
-
3X SSC spots have been observed to fluoresce by us and other microarray facilities for reasons still under investigation. However, it varies from batch to batch and grid to grid. Signals observed on the 3X SSC spots do not infer contamination of clones or carry-over during printing from neighbouring spots because black or base-level signals are constantly seen next to high-intensity spots.
- I see signals on the Arabidopsis spots when no RNA from Arabidopsis was added to the labelling reaction.
-
This is caused by non-specific binding of the labelled probe on the Arabidopsis spots. To prevent non-specific hybridisation, be sure to follow the recommended hybridisation and washing protocols (hybridise overnight with DIG Easy Hyb at 37°C for 16-18 hours and wash in 1x SSC/0.1% SDS at 50°C for 3 sets of 15 minutes, agitating twice during each set, followed by a thorough rinse in 1x SSC or 0.1x SSC at room temperature).
Other previously reported problems: